Ai Digital Biology
Bio Design
Biopharma Solutions Tools Tech
Reading Writing And Editing Dna
The 1-in-a-Million Breakthrough Behind Genyro’s Vision for a Post-Darwinian Age
It has never been easier to design new biology. Building it into DNA is still the bottleneck. Ahead of their appearance at SynBioBeta 2026 this May in San Jose, we spoke with Genyro co-founders Caltech Professor Kaihang Wang (Chief Scientific Advisor) and Adrian Woolfson (President and CEO) about the Wang Lab’s new Nature publication introducing Sidewinder, a major advance in DNA synthesis, and its role in Genyro’s broader vision for artificial biological intelligence.
From left to right: Genyro Co-founder Noah Robinson; Co-founder & Chief Scientific Advisor Professor Kaihang Wang; Co-founder, President & CEO Adrian Woolfson; Co-founder Professor Brian Hie. Photo credit: Marcus Ubungen.
Biology will not become truly programmable until DNA synthesis stops being the limiting step. Writing long, complex DNA, especially sequences rich in repeats or extreme GC content, remains slow, inaccurate, and expensive.
“Half the human genome is complex, repetitive, or GC-rich, and those regions are almost impossible to build,” explains Woolfson. “In plants, it’s closer to 85 percent. It’s a huge region of sequence space that you simply can’t navigate it, because you can’t construct it.”
The issue comes from how DNA is traditionally assembled. Most methods rely on complementary overlaps at fragment ends to stitch sequences together. Those overlaps serve two roles at once: they guide assembly, and they become part of the final DNA sequence. That coupling, Professor Kaihang Wang argues, is the root of the field’s long-standing accuracy ceiling.
“I have been suffering from the pain of DNA construction for decades,” Professor Kaihang Wang says. “When assembly instructions are coupled to the sequence itself through two-strand overlaps, you fundamentally cap specificity at about 90 to 97 percent, meaning misconnection rates as high as one in ten to one in thirty.”
In the lab’s new Nature paper, their Sidewinder method breaks that coupling between sequence and assembly instructions1. In Sidewinder, fragments locate their correct neighbors using barcodes that ligate at a three-way junction. After assembly, those barcodes are removed or displaced. The barcodes can be thought of as page numbers in a book, and the fragments as the information in the pages. The result is a seamless DNA sequence assembled with orders of magnitude higher accuracy than conventional approaches.
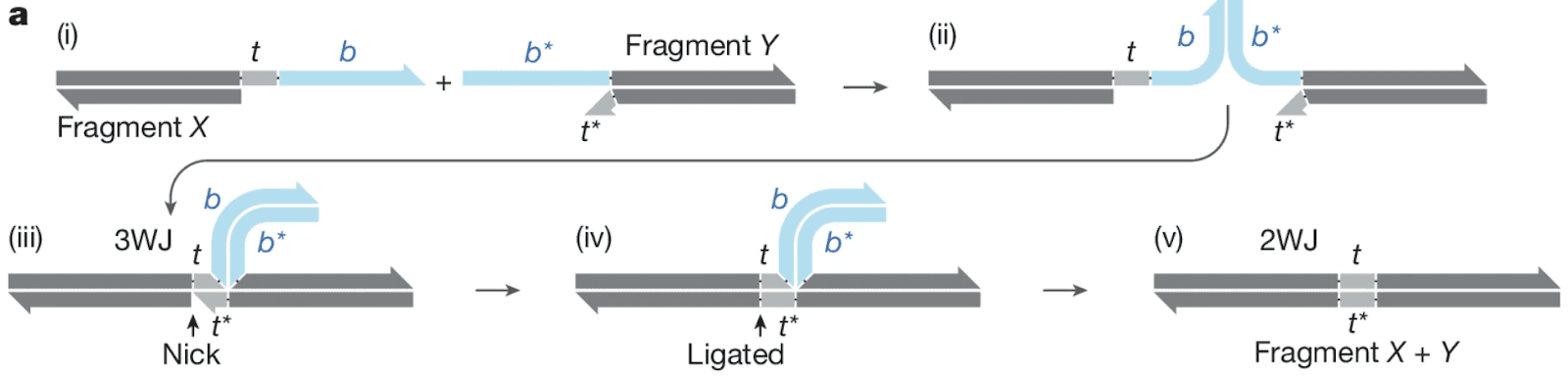
Figure 1a, Sidewinder directs DNA assembly through the formation of the 3 Way Junction. Sidewinder fragments contain short toehold pairs (t/t*) and long, unique barcode pairs (b/b*) (i). Sidewinder DNA assembly is directed by b/b* (ii). The association of b/b* brings together the two fragments, which are further stabilized by t/t* to form the 3WJ (iii). Toehold t* is ligated to the neighbouring fragment X, irreversibly connecting the two fragments (iv). The barcode helix b/b* is removed to restore the 2WJ, resulting in a scarless assembly (v).
The performance is not incremental. They report a misconnection rate of roughly 1 in 1 million, compared to a misconnection rate of 1 in 10 for previous methods. The team demonstrates reliable assembly across scenarios that routinely break in other methods, including 40-fragment constructs, high-GC and highly repetitive sequences, one-pot parallel assembly of multiple genes, and large combinatorial libraries comprising more than 440,000 variants with over 90% coverage.
The idea itself came in a moment familiar to many scientists. “I first saw this idea when I was taking a shower, and I instantly felt it was going to work,” Professor Kaihang Wang recalls. “But it would take a brave and brilliant PhD student, crazy and daring enough to realize what looked like an impossible design. I was very lucky to have Noah.”
That student, Noah Robinson, is the first author on the Sidewinder paper in Nature and a cofounder and CTO of Genyro, the newly launched stealth company that will license Sidewinder. Genyro's scientific advisory board includes some of the most influential figures in modern biology and engineering, including Nobel laureates Professors Frances Arnold and Roger Kornberg, as well as Professor Bob Langer, co-founder of Moderna, and Professor Jason Chin, Director of the Oxford Generative Biology Institute.
For Adrian Woolfson, co-founder, President, and CEO of Genyro, an accomplished author and alumnus of the MRC Laboratory of Molecular Biology in Cambridge, UK, Sidewinder is more than just a DNA assembly method. It is a foundational tool for his vision of Artificial Biological Intelligence, a concept he explores in his new book On the Future of Species. Artificial Biological Intelligence envisions a biology that moves beyond trial-and-error experimentation toward predicting and designing organisms directly from their sequence. That vision, however, depends on whether the physical act of building DNA can become reliable enough to keep pace with in silico design.
On the design side at Genyro, it’s as good as it gets. The company is also co-founded by Stanford professor and Arc Institute investigator Brian Hie. His lab’s large genome language model, Evo2, demonstrated the first functional bacteriophage genomes generated entirely by AI2. Whole-genome design remains a young field, but its rapid progress is likely to accelerate further as the challenges in genome assembly continue to vanish.
“We’re fortunate at Genyro to have leaders from both sides of the DNA design and construction problem under one roof,” Woolfson says. “On the design side, we have one of the world’s leading genome language model experts, Brian Hie, and on the construction side, Kaihang and Noah have built this extraordinary DNA assembly technology. Bringing design and construction together gives us a real path toward turning biology into a programmable engineering material. That won’t happen overnight, but even today, it allows us to do extraordinary things.”
The near-term goal is a much-needed upgrade to DNA synthesis. The long-term ambition is a world where genomes can be reliably built, predicted, and designed.
Meet Adrian Woolfson and Kaihang Wang at SynBioBeta in San Jose, May 4-7, 2026. Get your tickets here.
Robinson, N.E., Zhang, W., Ghosh, R. et al. Construction of complex and diverse DNA sequences using DNA three-way junctions. Nature (2026). https://doi.org/10.1038/s41586-025-10006-0
Samuel H. King, Claudia L. Driscoll, et al. Generative design of novel bacteriophages with genome language models. bioRxiv (2025). doi: https://doi.org/10.1101/2025.09.12.675911